Last week’s JACS highlighted Morgan’s recent paper, “Activity-Related Microsecond Dynamics Revealed by Temperature-Jump Förster Resonance Energy Transfer Measurements on Thermophilic Alcohol Dehydrogenase.” The Spotlight summary, included below, underscores the uniqueness of the timescales we access in our lab and the overall importance of understanding enzyme dynamics in catalysis. Congrats on an outstanding paper, Morgan!
Making Sense of Protein Motion in Enzymes, Erika Gebel Berg (Ph.D.)
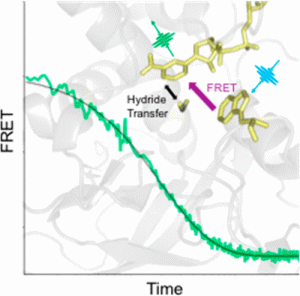
In recent years, scientists have come to appreciate that orchestrated protein motions are key drivers of enzyme activity. However, the details can be difficult to work out due to experimental limitations. Protein dynamics occur on multiple time scales, ranging from picoseconds to milliseconds, so measurement techniques must be carefully selected to probe the time scale of interest. Thomas Spiro, R. Brian Dyer, Judith Klinman, and colleagues use time-resolved temperature-jump FRET spectroscopy to probe the dynamics of thermophilic alcohol dehydrogenase, an enzyme model of the link between protein motion and activity (DOI: 10.1021/jacs.7b12369).
Previous researches in the Klinman Lab had indicated that protein motions facilitate the enzyme’s activity at temperature above 30 °C, but the timing and position of the motions was unknown. Now, by monitoring the energy transfer between NADH, the enzyme cofactor, and a nearby tryptophan side chain, the researchers observe a fast motion involving the enzyme active site that turns on above 30 °C. The fast relaxation rate is not detected at lower temperatures where the enzyme activity is compromised. This result implicates a link between thermally equilibrated, microsecond protein motions and the creation of active site configurations that facilitate catalysis. The work is a major step forward in understanding the functional role of enzyme dynamics.