How Do Different Organic Cover Crop Techniques Improve Soil Fertility and Impact Microbial Diversity?
Food Production
- Agriculture has been the foundation to civilization since the first human settlements, and in the wake of rapid population growth and awareness for organic farming, there has been a growing need for further investigation on the role of microbial presence in the soil and in the process of plant production.
- There are two main types of agriculture that play a large role providing the food that we need, conventional and organic. Compared to conventional farming, organic farming has a much more limited selection of tools and techniques that they can choose.
- One land management technique that both types of farming uses includes cover crops, which is a species of plant that is planted to protect and enrich the soil in preparation for cash crops. This is the primary technique that we will be focusing on for our research this summer.
- One role that cover crops play in increasing the fertility of soil is by introducing and fostering the growth of nitrogen-fixing bacteria that are responsible for providing nitrogen to the plants.
- Daniel Parson and students at the Oxford College Organic Farm – Source: http://news.emory.edu/stories/2015/10/er_best_college_farm/thumbs/story_main.jpg
Microbial Communities in the Soil
- An integral part of preparing the soil for cash crops is developing the microbial biota in the soil. Not only are they responsible for the cycling of organic matter and the generation of nutrients that the plants require, studies have shown that nitrogen-fixing rhizobia bacteria can determine plant community structure (Heijden et al. 2006).
- Symbiotic interactions between plants and bacterial communities are an integral part of the survival and success of both communities. In grassland communities with rhizobia communities in the soil, the plant productivity increased by 35%, the nitrogen content in the soil increased by 85% , and the community experienced a significant 34% increase in evenness (Marcel et al. 2006).
- One factor that influences the activity and community structure of soil microbial communities includes decomposing plant litter and the species of plants that are producing such litter (Bardgett and Shine 1999).
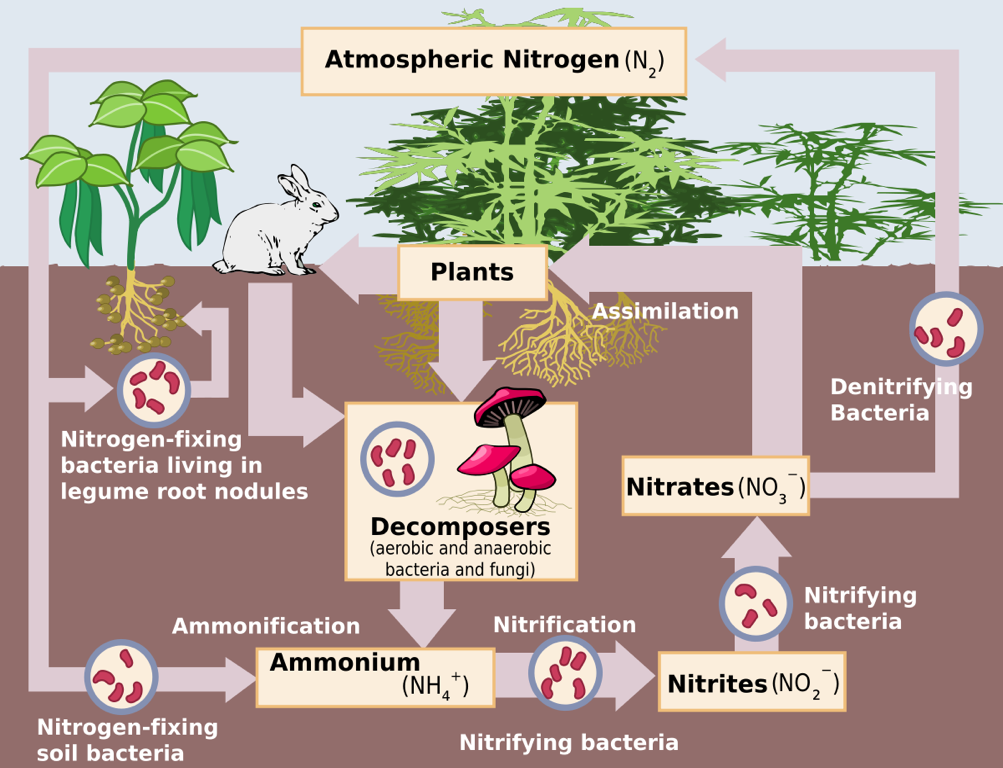
The Nitrogen Cycle – Source: https://upload.wikimedia.org/wikipedia/commons/thumb/f/fe/Nitrogen_Cycle.svg/1200px-Nitrogen_Cycle.svg.png
16 rRNA in the Bacterial Cell
- The bacterial cell is mainly composed of a capsule, cell wall, plasma membrane, circular DNA, cytoplasm, and ribosomes.
- The ribosomes in a bacterial cell functions in the production of proteins, an integral tool to the survival and reproduction of the cell.
- The ribosome is composed of a large subunit and a small subunit. Each subunit has rRNA that makes up part of the structure, one such rRNA in the small subunit is called the 16S rRNA.
- This rRNA strand is integral to the structure and function of the small subunit and thus it is high conserved throughout evolution.
- The 16s rDNA sequence is about 1500 base pairs long and there are specific regions within this sequence that are identical within a specie.
- These differences in these regions can be used to identify specific bacteria that is being studied, such as genus or species
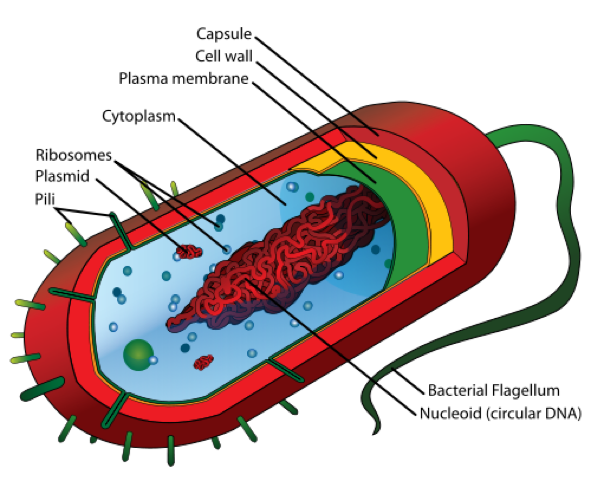
Structure of a Bacterial Cell – Source: https://upload.wikimedia.org/wikipedia/commons/thumb/5/5a/Average_prokaryote_cell-_en.svg/500px-Average_prokaryote_cell-_en.svg.pn
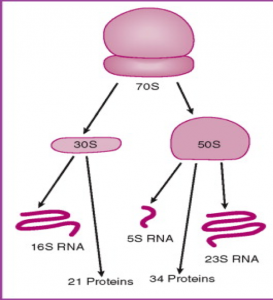
Bacterial Ribosomal Structure –
Source: http://ars.els-cdn.com/content/image/3-s2.0-B9780323034104500237-f17-10-9780323034104.jpg?httpAccept=%2A%2F%2A
16s rDNA Sequencing
- By amplifying the 16S rDNA of the bacterial cells, we will be able to analyze the genus and species of bacteria that this DNA came from.
- To analyze this piece of DNA, the first step is to amplify it through PCR. By conducting PCR, one copy of the 16S rDNA gene will be copied over and over again to where we will end with millions of the same copy of 16S rDNA.
- Then through purifying these copies of rDNA from the other proteins and molecules, we are able to retain a solution of many copies of the same 16S rDNA for sequencing.
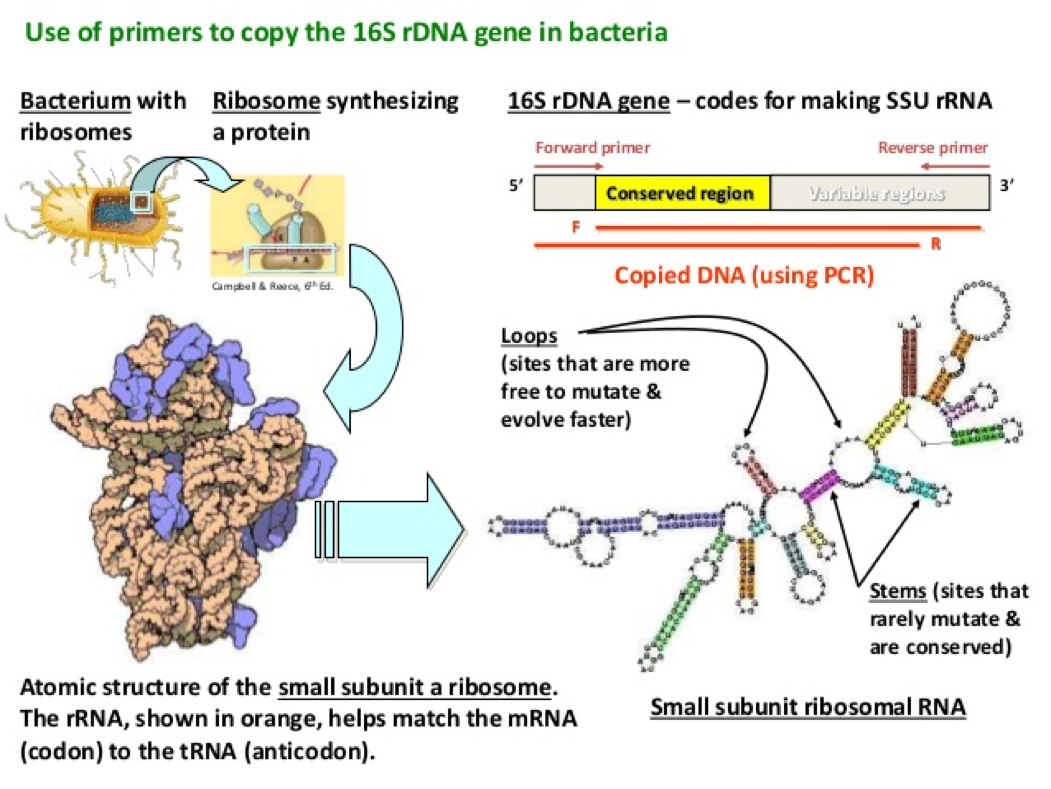
Source: https://image.slidesharecdn.com/16sribosomaldnasequenceanalysis-141130015317-conversion-gate01/95/16s-ribosomal-dna-sequence-analysis-13-638.jpg?cb=1417313002
Our Research
- This summer, we are focusing on the development of microbial diversity on the Oxford College Organic Farm through specific cover crop species and mixtures.
- To investigate this, we have taken an experimental plot and divided it into 9 sections, each section representing either a lone species of cover crop, a mega mix of 8 different species, or no cover crop.
- To observe change over time, we are collecting samples at 3 different time points, one is right after the cover crops are planted, another is right before the cover crops are mowed, and the last one is right after the cash crops are grown.
- In order to observe bacterial diversity, we are conducting DNA isolation and next-generation sequencing to amplify and identify bacterial species based on the 16s rDNA region.
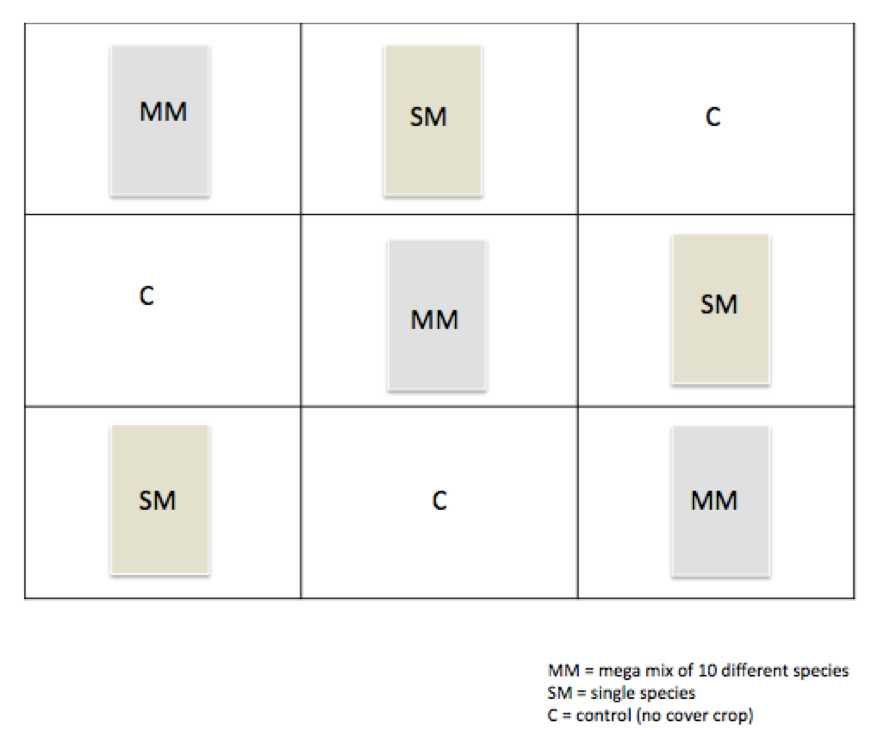
Experimental Plot Outline
- The method that we are taking is a thorough analysis of all the bacterial species in a soil sample. Since not all soil microbes can be cultured on a plate, this method is to give us an idea of the total microbial diversity in a sample.
- To achieve this, we will be isolating all of the DNA from a soil sample, which includes bacterial, fungal, eukaryotic, and all other DNA of organisms that may be present in the sample. Upon isolating these DNA, we will send them off to a lab where they will use specific primers that binds the variable regions 3 and 4 of the 16S gene and conduct PCR to amplify these bacterial DNA and to send off for sequencing using lasers.
- Upon receiving these results, we will analyze the specific species of each distinctive bacterial DNA sequence and then collecting these results in a spreadsheet to measure bacterial diversity.
- This type of sequencing is called Next-Generation Sequencing and the specific lab that we are sending our results to is using Illumina Sequencing Technology. By using this technology, we are able to sequence all of the different species of bacteria that are within a sample without having to go through culturing or colony isolation.
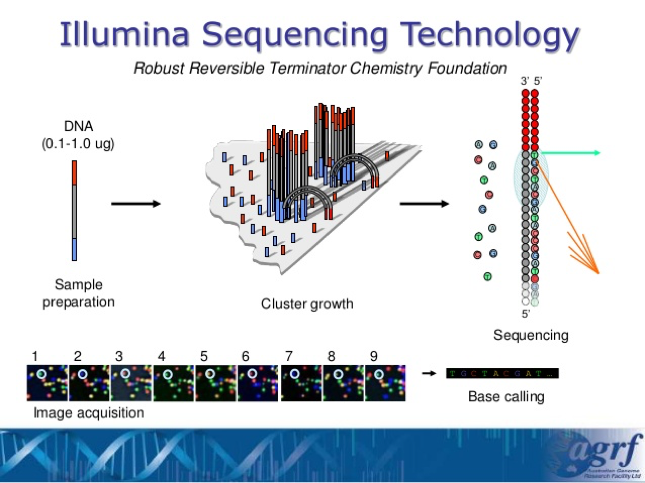
Next Generation Sequencing –
Source: https://image.slidesharecdn.com/kenmcgrath-nextgensequencing-gameofthronesedition-140707234700-phpapp01/95/ken-mcgrath-next-gen-sequencing-game-of-thrones-edition-8-638.jpg?cb=1405473639
Cover Crop Species and Characteristics
- The cover crop species that we use are shown in this diagram. The single species crop that will be used is buckwheat.

Selected Summer Cover Crop Species and Characteristics.
Measuring Bacterial Diversity
- Based on the results that we gather from next-generation sequencing, we will have a large database of all of the bacterial species that are present in all of the sections in our experimental plot as well as a potential track of changes in microbial diversity over time.
- By observing the appearance of new microbial species or disappearance of old microbial species in these plots over time, we can determine how the bacterial diversity of the experimental plot changed over time.
- Using the data we collect on these species, we will conduct literature search into these species to observe their specific effects of plant productivity. Depending on the nature of these bacterial species, they will demonstrate how the growth of cover crops can or cannot benefit future cash crop production.

Soil Microbial Diversity –
Source: https://cdn-asset-mel-1.airsquare.com/emnz/library/microbial-soil.jpg?201502270209
Our Research Thus Far
- Due to weather conditions, the experimental plot has been too wet to start planting cover crops, so we are currently actively optimizing all of the laboratory procedures that will take place once official sampling starts.
- To ensure that the samples we are collecting has bacteria that can be cultured and used in PCR , we plated some soil samples. The abundance of colonies grown these plates indicates that there are diverse bacterial species available to work with.
- After gathering samples from the experimental plot, we isolated the DNA from the samples and ran a PCR reaction with these isolated DNA. We ran a gel electrophoresis with the results of this PCR reaction to show that there was indeed bacterial DNA in the soil sample isolated and they were able to be amplified and thus sequenced.
- To ensure the purity of these isolated DNA, we also read the concentrations of each sample. The highest concentrations of these samples were 92.2 ng/uL and 23.9 ng/uL respectively and the 260:280 ratios demonstrated strong purity within both samples.
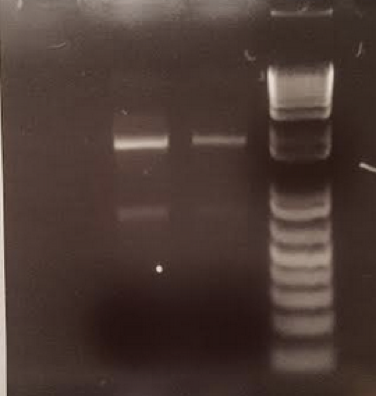
Gel Electrophoresis Results
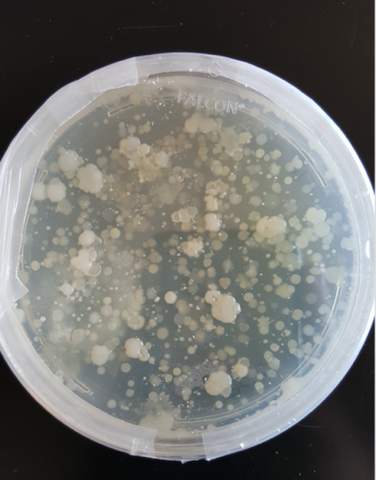
Bacterial Colonies from Soil Samples
Future Directions
- Our next steps include tilling the beds on the experimental plot and planting the cover crop seeds. After a week of planting cover crop seeds, we will take our first samples to start isolation and prepare for sequencing.
- We will also be finding methods for determining soil fertility by measuring organic matter content and nitrogen/carbon content.
- After we have ensured that all of our samples have enough DNA concentration, we will be sending all of the results off at once to the UGA lab where the Illumina Sequencing Technology will take place and help analyze these DNA.
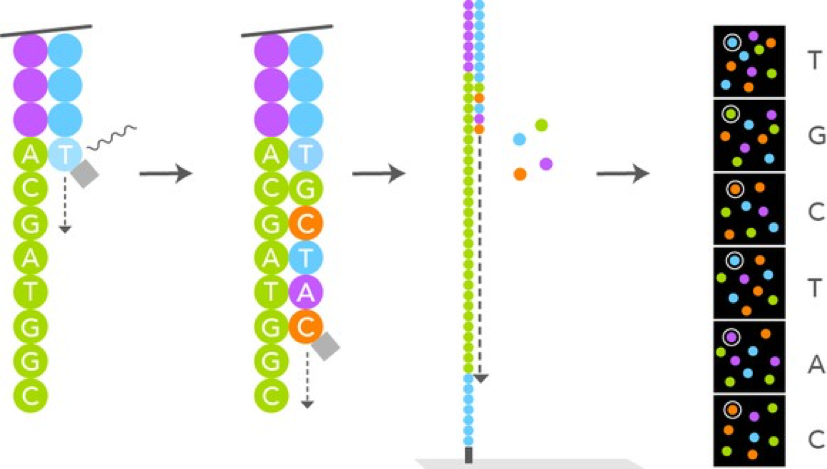
Illumina Sequencing Analysis –
Source: https://g.foolcdn.com/editorial/images/433008/hires-sbs-sequencing_large.jpg
Sources Cited
- Heijden, Van Der, Marcel G.a, Roy Bakker, Joost Verwaal, Tanja R. Scheublin, Matthy Rutten, Richard Van Logtestijn, and Christian Staehelin. “Symbiotic Bacteria as a Determinant of Plant Community Structure and Plant Productivity in Dune Grassland.” FEMS Microbiology Ecology 56, no. 2 (May 1, 2006): 178–87.
- Bardgett, Richard D., and Amanda Shine. “Linkages between Plant Litter Diversity, Soil Microbial Biomass and Ecosystem Function in Temperate Grasslands.” Soil Biology and Biochemistry 31, no. 2 (February 1, 1999): 317–21. doi:10.1016/S0038-0717(98)00121-7.
- Marcel G.A. Van Der Heijden, Bakker, R., Verwaal, J., Scheublin, T., Rutten, M., Van Logtestijn, R., and Staehelin, C. “Symbiotic bacteria as a determinant of plant community structure and plant productivity in dune grassland” FEMS Microbiology Ecology, 2006, vol. 56, pp. 178- 187.

Source: https://pics.me.me/allbacteriais-bad-false-bacteria-is-necessary-in-the-carbon-anonitrogencycle-16197788.png

thank you good post. like like. like