 Understanding how eukaryotic pathogens generate genetic and phenotypic variation has meaningful clinical and evolutionary implications. The Hickman lab investigates the genome dynamics of the yeast Candida albicans, a commensal that primarily resides in human gastrointestinal tracts and yet causes a wide-range of infections, depending on host context. Exposure to stressors often elevates genome instability and the resulting increased genetic provides the underpinnings for natural selection to facilitate adaption. A particularly relevant adaptive trait is antifungal drug resistance, a phenomenon that predominantly occurs de novo during cell division. We are interested in characterizing the environmental drivers and cellular mechanisms resulting in genomic alterations, ultimately leading to fungal adaptation.
Understanding how eukaryotic pathogens generate genetic and phenotypic variation has meaningful clinical and evolutionary implications. The Hickman lab investigates the genome dynamics of the yeast Candida albicans, a commensal that primarily resides in human gastrointestinal tracts and yet causes a wide-range of infections, depending on host context. Exposure to stressors often elevates genome instability and the resulting increased genetic provides the underpinnings for natural selection to facilitate adaption. A particularly relevant adaptive trait is antifungal drug resistance, a phenomenon that predominantly occurs de novo during cell division. We are interested in characterizing the environmental drivers and cellular mechanisms resulting in genomic alterations, ultimately leading to fungal adaptation.
Causes and consequences of genome plasticity
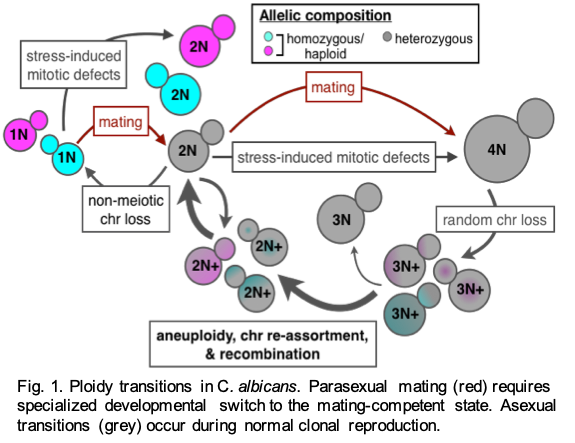
One of our major, long-standing lines of inquiry is to understand how fungi manipulate their ploidy (i.e. the number of sets of chromosomes) to drive genetic variation. Shifts in ploidy are mediated by both conventional sexual cycles as well as asexual mechanisms promote genetic diversity within a population of cells. Ploidy transitions facilitate large-scale genome perturbations, including recombination, aneuploidy and allelic re-assortment and can occur within a single cell division. While C. albicans was long considered to be an obligate diploid organism, we discovered a viable haploid stage that C. albicans uses to purge its genome of deleterious recessive mutations that accumulate in the diploid state (Hickman et al 2013 Nature). Polyploid C. albicans are also viable and can arise via mating or mitotic defects and have been isolated from patient samples and in some cases are highly resistant to antifungal drugs.
These dramatic deviations from diploidy are often transient, suggesting a selective force acts to return C. albicans to the diploid state, a phenomenon that we term ‘ploidy drive’ (Hickman et al 2015 Genetics; Gerstein et al 2017 Evolution). We are currently investigating how mutation rates differ among C. albicans ploidy states, how exposure to antifungal drugs differentially induces mutagenesis depending on ploidy, and how these interactions potentially drive the rapid emergence of antifungal drug resistance. Additional research questions we are pursuing include determining the molecular mechanisms of chromosome and ploidy instability, identifying abiotic and host environments that promote ploidy transitions, and assessing the functional consequences and phenotypic diversity generated through these processes.
Host-microbe interactions and the evolution of fungal pathogens
The Hickman lab is also addressing how host-microbe interactions serve to drive virulence phenotypes, pathogen genome plasticity, and ultimately evolution of fungal pathogens. As an opportunistic pathogen, C. albicans resides within hosts for significant periods of time without ever causing harm, yet are capable of infection. Infection dynamics are the result of fungal strain background in conjunction with host genotype and environment. Susceptible host contexts include immunocompromised individuals or healthy hosts taking antibiotics, and thus clearing their bacterial microbiomes. We have recently adopted the nematode Caenorhabditis elegans as an experimental host system to study the genome dynamics of fungal colonization, infection and evolution. We have found that infection with C. albicans significantly reduces multiple measures of host fitness, including survival and reproduction, and we have developed a novel assay that rapidly assesses overall host fitness that can be used to screen virulence phenotypes of different pathogen genetic backgrounds, other yeast species, and host genotypes in a highly quantitative manner (Fiestel et al 2019). We are leveraging our C. elegans – C. albicans host-microbe experimental system to perform both genetic screens and experimental evolution studies to identify how host and pathogen genetic backgrounds interact to produce virulent phenotypes. Additional research questions we are pursuing include determining how host immune function and/or the host polymicrobial communities impact fungal genome plasticity and how these genomic alterations contribute to virulence evolution and resistance to antifungal drugs.
Funding:
2020-2024 NSF CAREER (DEB-1943415) – Determining the impact of asexual ploidy transitions on evolutionary trajectories.
Award abstract: Genetic variation is important for species to be able to survive and adapt to changing environments. For most plants and animals, sex is used to generate variation and involves both mating and meiosis. Meiosis, the process by which a cell divides to produce four cells with half the genetic information, makes new genetic combinations as whole chromosomes and parts of chromosomes reshuffle during the division process. However, for many fungi, sexual cycles are either absent, rarely occur or substantially deviate from conventional mating and meiosis systems. Instead, fungi, including many species that commonly infect plants and/or animals, can change the number of chromosomes and chromosome sets (ploidy) they contain during asexual growth. Notably, there is tremendous ploidy variation among strains of fungal pathogens that interact with host environments and encounter antifungal drugs used to treat infected hosts. This research leverages Candida albicans, an opportunistic fungal pathogen of humans, as an experimental system to identify how frequently changes in ploidy occur, and what environments promote these changes. We will also study how fungal pathogens’ capacity to change ploidy influence the evolution of virulence and antifungal drug resistance. As part of our mission to undergraduate science education and literacy, we will develop an upper-division writing-intensive class designed to teach students how to transform experimental results from data to knowledge while challenging students to distill scientific knowledge into a framework accessible to the general public.
While asexual ploidy transitions are well-documented to occur in many fungal species and across diverse growth environments, whether they drive evolutionary processes or are simply a consequence of elevated genome instability remains to be explicitly tested experimentally. Furthermore, the environmental signals and pathways to ploidy changes have not been well characterized, nor have the consequences of asexual ploidy transitions been tested within an evolutionary framework. By screening for Candida genome size changes across many in vitro and host environments at short- and mid-length time scales, environments that promote ploidy transitions will be identified. Then, through constructing C. albicans strains that are genetically-locked into the diploid or polyploid state, the researchers will test how ploidy transitions contribute to evolutionary processes through experimental evolution experiments and compare the rates and outcomes following selection for virulence and antifungal drug resistance. This research program uses C. albicans ploidy variation as a compelling biological context for undergraduate STEM education. I will develop a science-writing course that utilizes data generated in an introductory biology lab module that measures mutation rates across C. albicans ploidy states. The educational mission of this proposal synergizes with the broader impacts associated with this research, which will reveal important insights into how fungal pathogens evolve virulence and drug resistance.