Overview
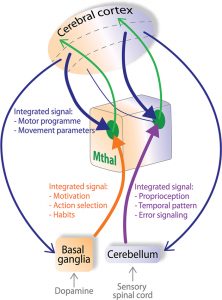
The basal ganglia and the cerebellum are intimately connected with the cerebral cortex through the thalamus, such that together these brain area are involved in movement control and cognition.
Dysfunction of this network of brain structure results in motor disorders, such as ataxia when the cerebellum is impaired and Parkinson’s disease when the dopamine innervation of the basal ganglia degenerates.
Techniques we use
-
Mouse Behavior
-
Optogenetics
-
Cortical Voltage Imaging
-
Electrophysiology in vivo
-
Electrophysiology in vitro
-
Computer Modeling
-
Histology
Specific Aims and Experimental Approach
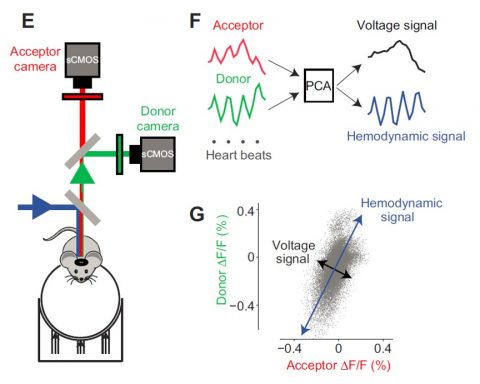
1) To study cortical motor processing we employ widefield cortical voltage imaging of specific cortical cell types during the acquisition and performance of cued lick- and reaching tasks in head-fixed mice. This allows us to determine behaviorally relevant cortical activity patterns through dimensionality reduction methods and event aligned activity traces in regions of interest.
2) A major project in the lab is concerned with alterations in cortical processing in Parkinsonian conditions. We use the progressive MitoPark mouse model to determine the time course of cortical dysfunction with progressive motor disability through voltage imaging and electrophysiology.
Computer Modeling
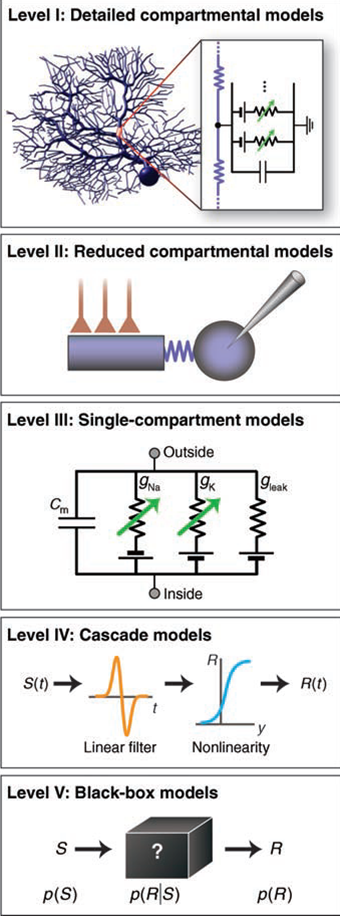
The underlying strategy in the lab is to combine computer modeling and experimental investigations of neural properties to address the question of how computation in specific brain circuits might occur.
We construct detailed compartmental single neuron models of motor thalamic and motor cortical L5 neurons using the NEURON software. This work accompanies our
experimental efforts to allow a mechanistic understanding of how excitability changes affect synaptic integration.
To build a single neuron model we first obtain whole cell recordings from rat brain slices to characterize basic properties the model needs to show. Neurons are visualized with infrared optics using a water immersion objective. We use glass pipettes to obtain intracellular electrical recordings from visualized cells in the whole cell configuration. Different experimental manipulations we use include application of specific pharmacological blockers to isolate specific voltage-gated currents in our neurons, and electrical stimulation of input pathways to examine synaptic properties. Most important, we use the technique of dynamic current clamping to apply artificial synaptic inputs in vitro. This technique allows us to study the input/output processing of neurons in controlled conditions, while we mimic the input condition as it might occur in the behaving animal. Synaptic conductance waveforms are typically derived from computational models and dynamic current clamping allows us to directly compare model neurons and neurons recorded in vitro.
|