Please see Katia’s google scholar page for more recent publications!
2021
Braun, K.M., Moreno, G.K., Halfmann, P.J., Hodcroft, E.B., Baker, D.A., Boehm, E.C., Weiler, A.M., Haj, A.K., Hatta, M., Chiba, S. Maemura, T., Kawaoka, Y., Koelle, K., O’Connor, D.H., Friedrich, T.C. (2021). Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. *authors contributed equally. bioRxiv. doi: https://doi.org/10.1101/2020.11.16.384917
2020
Gallagher, M.E., Sieben, A.J., Nelson, K.N., Kraay, A.N.M., Orenstein, W.A., Lopman, B., Handel, A., Koelle, K. (2020). Indirect benefits are a crucial consideration when evaluating SARS-CoV-2 vaccine candidates. Nature Medicine. doi: https://doi.org/10.1038/s41591-020-01172-x
 Moreno, G.K.*, Braun K.M.*, Riemersma, K.K.*, Martin, M.A., Halfman, P.J., Crooks, C.M., Prall, T., Baker, D., Baczenas, J.J., Heffron, A.S., Ramuta, M., Khubbar, M., Weiler, A.M., Accola, M.A., Rehrauer, W.M., O’Connor, S.L., Safdar, N., Pepperell, C.S., Dasu, T., Bhattacharyya, S., Kawaoka, Y., Koelle, K., O’Connor, D.H., Friedrich, T.C. (2020). *authors contributed equally. Revealing fine-scale spatiotemporal differences in SARS-CoV-2 introduction and spread. Nature Communications. doi: https://doi.org/10.1038/s41467-020-19346-z
Moreno, G.K.*, Braun K.M.*, Riemersma, K.K.*, Martin, M.A., Halfman, P.J., Crooks, C.M., Prall, T., Baker, D., Baczenas, J.J., Heffron, A.S., Ramuta, M., Khubbar, M., Weiler, A.M., Accola, M.A., Rehrauer, W.M., O’Connor, S.L., Safdar, N., Pepperell, C.S., Dasu, T., Bhattacharyya, S., Kawaoka, Y., Koelle, K., O’Connor, D.H., Friedrich, T.C. (2020). *authors contributed equally. Revealing fine-scale spatiotemporal differences in SARS-CoV-2 introduction and spread. Nature Communications. doi: https://doi.org/10.1038/s41467-020-19346-z
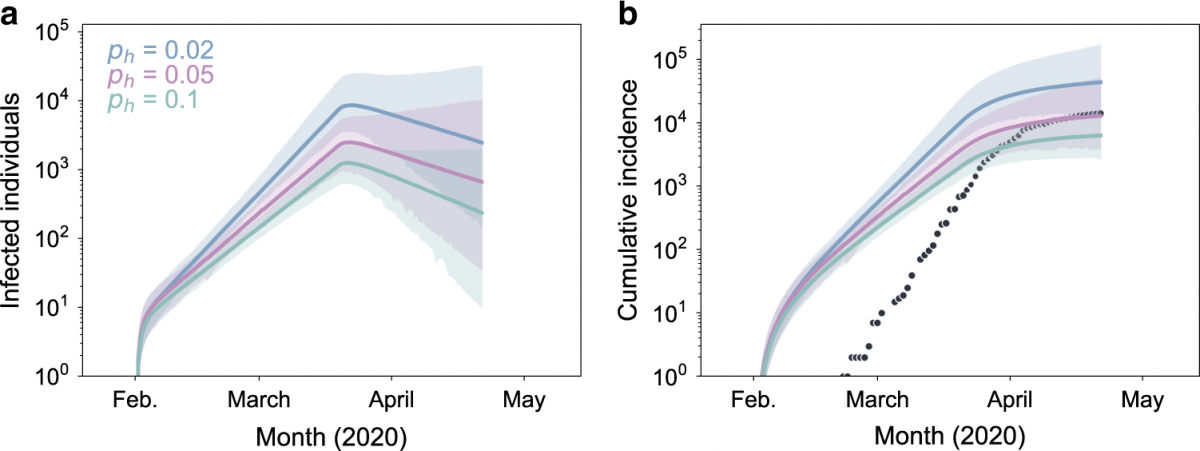
Miller, D.*, Martin, M.A.*, Harel, N.*, Tirosh, O.*, Kustin, T.*, Meir, M., Sorek, N., Gefen-Halevi, S., Amit, S., Vorontsov, O., Shaag, A., Wolf, D., Peretz, A., Shemer-Avni, Y., Roif-Kaminsky, D., Kopelman, N.M., Huppert, A., Koelle, K., Stern, A. (2020). Full genome viral sequences inform patterns of SARS-CoV-2 spread into and within Israel. *authors contributed equally. Nature Communications. doi: https://doi.org/10.1038/s41467-020-19248-0
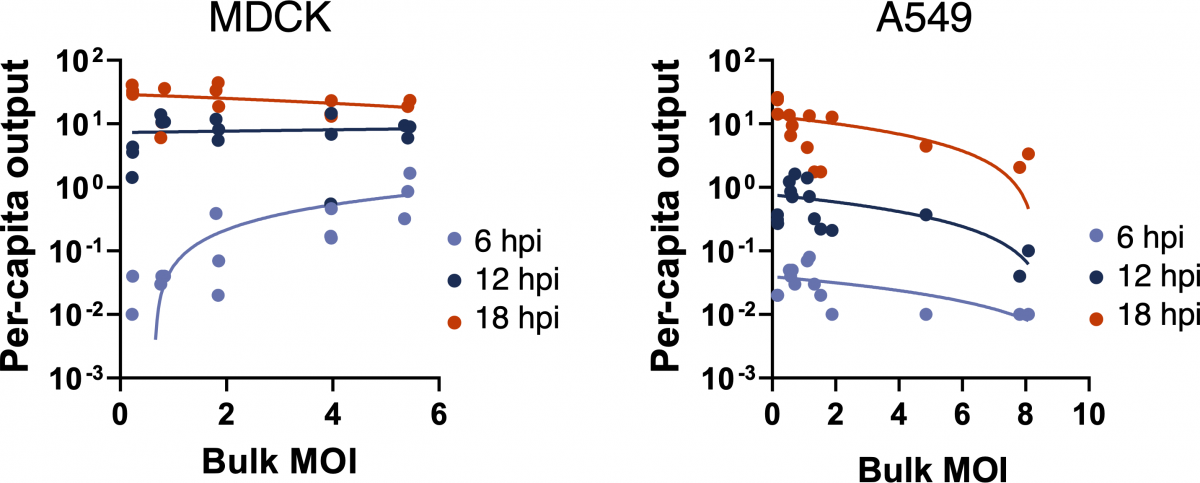
Martin, B.E.*, Harris, J.D.*, Sun, J., Koelle, K., Brooke, C.B. (2020). Cellular co-infection can modulate the efficiency of influenza A virus production and shape the interferon response. *authors contributed equally. PLoS Pathogens. doi: https://doi.org/10.1101/752329
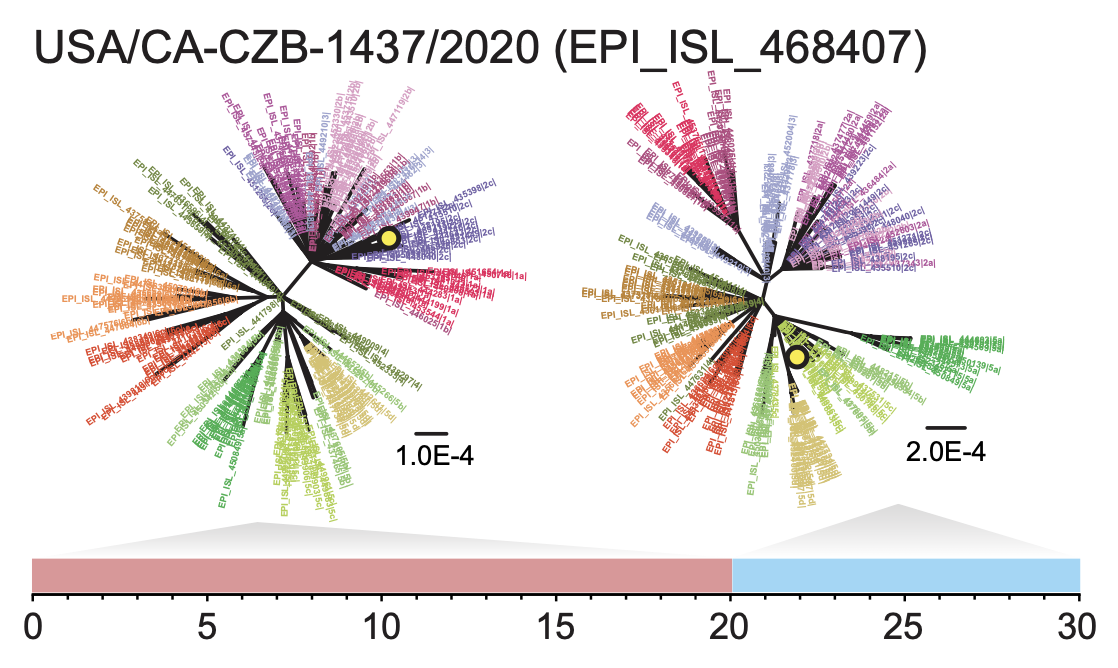
VanInsberghe, D., Neish, A., Lowen, A.C., Koelle, K. (2020). Identification of SARS-CoV-2 recombinant genomes. bioRxiv. doi: https://doi.org/10.1101/2020.08.05.238386
Steele, M.K., Wikswo, M.E., Hall, A.J., Koelle, K., Handel, A., B.A. Lopman (2020). Characterizing norovirus transmission from outbreak data in the United States. Emerg Infect Dis. doi: https://dx.doi.org/10.3201/eid2608.191537
Caudill, V., …[Koelle, K. author 10 of 51]. and P.S. Pennings (2020). CpG-creating mutations are costly in many human viruses. Evolutionary Ecology. doi: https://doi.org/10.1007/s10682-020-10039-z
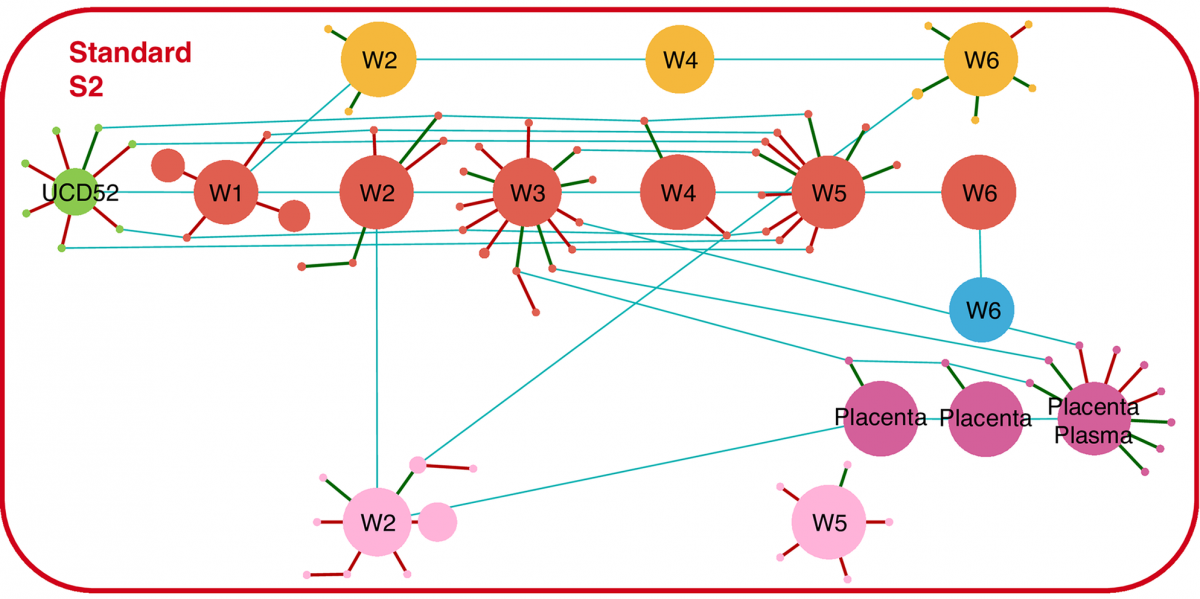
Vera Cruz, D., Nelson, C.S., Tran, D., Barry, P.A., Kaur, A., Koelle, K.*, S. Permar* (2020). Intrahost cytomegalovirus population genetics following antibody pretreatment in a monkey model of congenital transmission. *authors contributed equally. PLoS Pathogens. doi: https://doi.org/10.1371/journal.ppat.1007968
Martin M.A., Woods, C.W., Koelle, K. (in revision). The Dynamics of Influenza A H3N2 Defective Viral Genomes from a Human Challenge Study. bioRxiv. doi: 10.1101/814673
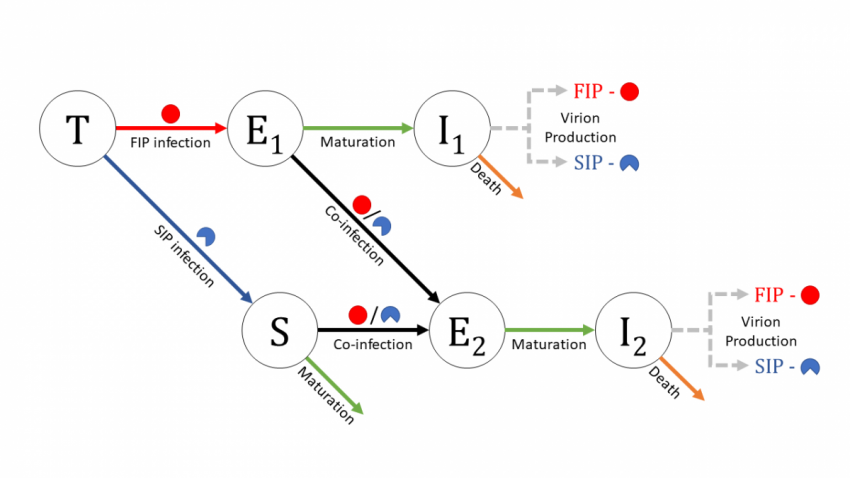
Farrell, A., Brooke, C., Koelle, K., and R. Ke (in review). Coinfection of semi-infectious particles can contribute substantially to influenza infection dynamics. bioRxiv. doi:10.1101/547349
2019
Craig, R., Kunkel, E., Crowcroft, N., Fitzpatrick, M., de Melker, H., Althouse, B., Merkel, T., Scarpino, S.V., Koelle, K., Carlsson, R., and S. Bolotin (2019). Asymptomatic infection and transmission of pertussis in households: A systematic review. Clinical Infectious Diseases, doi: 10.1093/cid/ciz531
Koelle, K., Farrell, A., Brooke, C., and R. Ke (2019) Within-host infectious disease models accommodating cellular coinfection, with an application to influenza. Virus Evolution. doi: 10.1093/ve/vez018
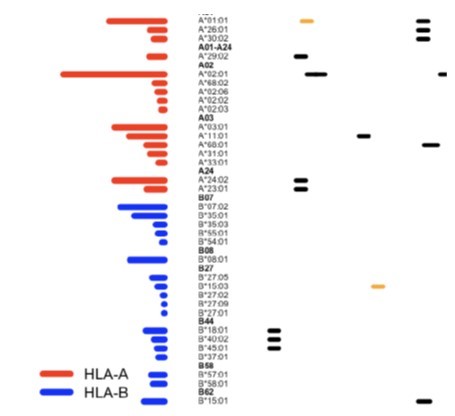
Li, Z., Zarnitsyna, V., Lowen, A., Weissman, D., Koelle, K., Kohlmeier, J. and R. Antia (2019). Why Are CD8 T Cell Epitopes of Human Influenza A Virus Conserved? Journal of Virology. doi: 10.1128/JVI.01534-18
2018
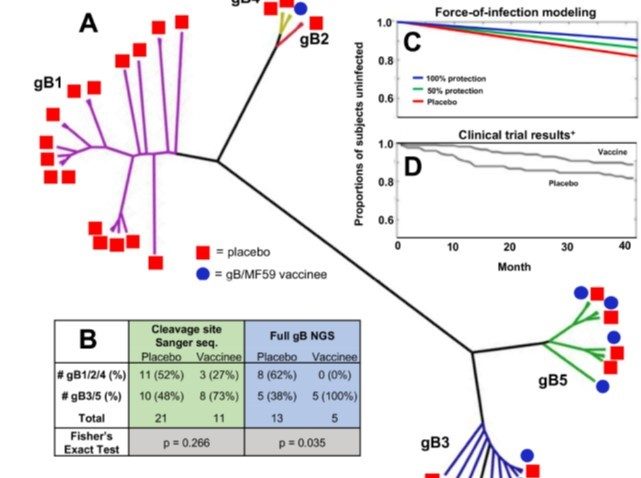
Nelson, C., Vera Cruz, D. Su, M., Xie, G., Vandergrift, N., Pass, R., Forman, M., Diener-West, M., Koelle, K., Arav-Boger, R. and S. Permar (2018). Intrahost dynamics of human cytomegalovirus variants acquired by seronegative glycoprotein B vaccinees. Journal of Virology. doi: 10.1128/JVI.01695-18
Gallagher, M. E., Brooke, C.B., Ke, R., and K. Koelle (2018). Causes and consequences of spatial within-host viral spread. Viruses. doi: 10.3390/v10110627.
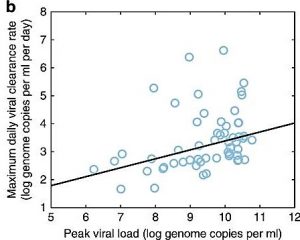
Ben-Shachar, R. and K. Koelle (2018). Transmission-clearance trade-offs indicate that dengue virulence evolution depends on epidemiological context. Nature Communications. doi: 10.1038/s41467-018-04595-w.
McKenney, E., Koelle, K., Dunn, R., and A. Yoder (2018). The ecosystem services of animal microbiomes. Molecular Ecology. doi: 10.1111/mec.14532
2017
Raghwani, J., Thompson, R., and K.Koelle (2017). Selection on non-antigenic gene segments of seasonal influenza A virus and its impact on adaptive evolution. Virus Evolution. doi: 10.1093/ve/vex034
Sobel Leonard, A., Weissman, D., Greenbaum, B., Ghedin, E., and K. Koelle (2017). Transmission bottleneck size estimation from pathogen deep-sequencing data, with an application to human influenza A virus. Journal of Virology. doi: 10.1128/JVI.00171-17
Nelson, C.S., Vera Cruz, D., Tran, D., Bialas, K.M., Stamper, L., Wu, H., Gilbert, M., Blair, R., Alvarez, X., Itell, H., Chen, M., Deshpande, A., Chiuppesi, F., Wussow, F., Diamond, D.J. Vandergrift, N., Walter, M., Barry, P.A., Cohen-Wolkowiez, M., Koelle, K., Kaur, A., and S.R. Permar (2017). Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight. doi: 10.1172/jci.insight.94002
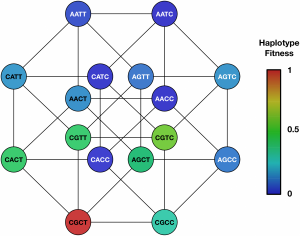
Sobel Leonard, A., McClain, M., Smith, G. Wentworth, D. Halpin, R., Lin, X., Ransier, A., Stockwell, T., Das, S., Gilbert, A., Lambkin-Williams, R., Ginsburg, G., Woods, C., Koelle, K., and C. Illingworth (2017). Limited effective reassortment shapes influenza evolution in a human host. PLoS Pathogens. doi: 10.1371/journal.ppat.1006203
2016
Flasche, S.*, Jit, M., Rodriguez-Barrauer*, I., Coudeville*, L.,Recker*, M., Koelle*, K., Milne*, G., Hladish*, T., Perkins*, A., Dorigatti, I., Cummings, D., Espana, G., Kelso, J., Longini, I., Lorenco, J., Pearson, C., Reiner, R.C., and N. Ferguson (2016) The long term safety, public health impact, and cost effectiveness of routine vaccination with Dengvaxia®: a model comparison study. PLoS Medicine. *authors contributed equally. doi: 10.1371/journal.pmed.1002181
Ben-Shachar, R., Schmidler, S.C., and K. Koelle (2016). Drivers of inter-individual variation in dengue viral load dynamics. PLoS Computational Biology. doi: 10.1371/journal.pcbi.1005194
Sobel Leonard, A., McClain, M., Smith, G. Wentworth, D. Halpin, R., Lin, X., Ransier, A., Stockwell, T., Das, S., Gilbert, A., Lambkin-Williams, R., Ginsburg, G., Woods, C., and K. Koelle (2016) Deep sequencing of influenza A virus from a human challenge study reveals a large founder population size and rapid intrahost viral evolution. Journal of Virology. Selected as a Journal of Virology Spotlight Feature. doi: 10.1128/JVI.01657-16
Flasche, S.*, Jit, M., Rodriguez-Barrauer*, I., Coudeville*, L., Recker*, M., Koelle*, K., Milne*, G., Hladish*, T., Perkins*, A., Dorigatti, I., Cummings, D., Espana, G., Kelso, J., Longini, I., Lorenco, J., Pearson, C., Reiner, R.C., and N. Ferguson (2016) Comparative modelling of dengue vaccine public health impact. Report to the Strategic Advisory Groups of Experts (SAGE) on Immunization. (SAGE recommendations will be the basis for WHO global public health policy for appropriate use of Dengvaxia®, Sanofi Pasteur’s dengue vaccine.) *authors contributed equally.
2015
Koelle, K. and D.A. Rasmussen (2015). The effects of a deleterious mutation load on patterns of influenza A/H3N2’s antigenic evolution in humans. eLife. doi: 10.7554/eLife.07361
Ben-Shachar, R. and K. Koelle (2015). Minimal within-host dengue models highlight the specific roles of the immune response in primary and secondary dengue infections. Journal of the Royal Society, Interface. 12 (103), p. 20140886. doi: 10.1098/rsif.2014.0886
2014
Koelle, K. and D.A. Rasmussen (2014). Influenza: prediction is worth a shot. Nature. 507 (7490), pp. 47-48. doi:10.1038/nature13054
Rasmussen, D.A., Volz, E., and K. Koelle (2014). Phylodynamic inference for structured epidemiological models. PLoS Computational Biology. 10(4), p. e1003570. doi: 10.1371/journal.pcbi.1003570
Rasmussen, D.A., Boni, M., and K. Koelle (2014). Reconciling phylodynamics with epidemiology: The case of dengue virus in southern Vietnam. Molecular Biology and Evolution. 31(2): pp.258-271. doi: 10.1093/molbev/mst203
2013
Scholle, S.O., Ypma, R., Lloyd, A.L., and K. Koelle (2013). Viral substitution rate variation can arise from the interplay between within-host and epidemiological dynamics. The American Naturalist. 182(4), pp.494-513. doi: 10.1086/672000. (Stacy received Honorable Mention for the 2013 Student Paper of the Year Award from the American Naturalist for this paper.)
Yuan, H. and K. Koelle (2013). The evolutionary dynamics of receptor binding avidity in influenza A: a mathematical model for a new antigenic drift hypothesis. Philosophical Transactions of the Royal Society, B. 368 (1614): p. 20120204. doi: 10.1098/rstb.2012.0204
Luo, S. and K. Koelle (2013). Navigating the devious course of evolution: the importance of mechanistic models for identifying eco-evolutionary dynamics in nature. The American Naturalist.181, pp. S58-S75. doi: 10.1086/669952
Volz, E.M., Koelle, K., and T. Bedford (2013). Viral phylodynamics. PLoS Computational Biology. 9(3): e1002947. doi: 10.1371/journal.pcbi.1002947
Wu, S., Koelle, K., and A. Rodrigo (2013). Coalescent entanglement and the conditional dependence of the times to common ancestry of mutually exclusive pairs of individuals. Journal of Heredity. 104 (1): 86-91. doi: 10.1093/jhered/ess074
2012
Ratmann, O., Donker, G., Meijer, A., Fraser, C., and K. Koelle (2012). Phylodynamic inference and model assessment with Approximate Bayesian Computation: influenza as a case study. PLoS Computational Biology. 8(12):e1002835. doi: 10.1371/journal.pcbi.1002835
Luo, S., Reed, M., Mattingly, J., and K. Koelle (2012). The impact of host immune status on the within-host and population dynamics of antigenic immune escape. Journal of the Royal Society, Interface. 9 (75), pp. 2603-2613. doi: 10.1098/rsif.2012.0180
Arinaminpathy, N., Ratmann, O., Koelle, K., Epstein, S.L., Price, G.E., Viboud, C., Miller, M., and B. Grenfell (2012). Impact of cross-protective vaccines on the epidemiological and evolutionary dynamics of influenza. PNAS. 109(8), pp. 3173-3177. doi: 10.1073/pnas.1113342109
WHO-VMI Dengue Vaccine Modeling Group (2012). Assessing the potential of a candidate dengue vaccine with mathematical modeling. PLoS Neglected Tropical Diseases. 6(3), e1450. doi: 10.1371/journal.pntd.0001450 (The group consists of 28 experts in dengue epidemiology, clinical practice, immunology, virology, vaccinology, entomology, and mathematical modeling.)
Koelle, K. and D.A. Rasmussen (2012). Rates of coalescence for common epidemiological models at equilibrium. Journal of the Royal Society, Interface. 9(70), pp.997-1007. doi: 10.1098/rsif.2011.0495
2011
Meyers, L.A., Kerr, B., and K. Koelle (2011). Network Perspectives on Infectious Disease Dynamics. Interdisciplinary Perspectives on Infectious Diseases. Volume 2011 (2011), Article ID 146765. doi: 10.1155/2011/146765
Rasmussen, D.A., Ratmann, O., and K. Koelle (2011). Inference for nonlinear epidemiological models using genealogies and time series. PLoS Computational Biology. 7 (8), e1002136. doi: 10.1371/journal.pcbi.1002136
Koelle, K., Ratmann, O., Rasmussen, D.A., Pasour, V., and J. Mattingly (2011). A dimensionless number for understanding the evolutionary dynamics of antigenically variable RNA viruses. Proceedings of the Royal Society, Series B. 278, pp. 3723-3730. doi: 10.1098/rspb.2011.0435
≤2010
Koelle, K., Khatri, P., Kamradt, M., and T. Kepler (2010). A two-tiered model for studying the ecological and evolutionary dynamics of rapidly evolving viruses, with an application to influenza. Journal of the Royal Society, Interface. 7, pp.1257-1274. doi: 10.1098/rsif.2010.0007
Koelle, K., Kamradt, M., and M. Pascual (2009). Understanding the dynamics of rapidly evolving pathogens through modeling the tempo of antigenic change: influenza as a case study. Epidemics. 1, pp. 129-137. doi: 10.1016/j.epidem.2009.05.003
Koelle, K (2009). The impact of climate on the disease dynamics of cholera. Clinical Microbiology and Infection. 15(s1), pp. 29-31. doi: 10.1111/j.1469-0691.2008.02686.x
Nagao, Y. and K. Koelle (2008). Decreases in dengue transmission may act to increase the incidence of Dengue Hemorrhagic Fever. PNAS. 105(6), pp. 2238-2243. doi: 10.1073/pnas.0709029105
Cobey, S. and K. Koelle (2008). Capturing escape in infectious disease dynamics. Trends in Ecology and Evolution. 23(10), pp.572-577. doi: 10.1016/j.tree.2008.06.008
Pascual, M., Cazelles, B., Bouma, M., Chaves, L., and K. Koelle (2008). Shifting patterns: malaria dynamics and rainfall variability in an African highland. Proceedings of the Royal Society, B. 275. pp.123-132.
Finkelman, B., Viboud, C., Koelle, K., Ferrari, M., Bharti, N., and B. Grenfell (2007). Global Patterns in Seasonal Activity of Influenza A/H3N2, A/H1N1, and B from 1997 to 2005: Viral Coexistence and Latitudinal Gradients. PLoS One. 2(12): e1296.
Koelle, K., Cobey, S., Grenfell, B. and M. Pascual (2006). Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 314. pp. 1898-1903.
Koelle, K., Pascual, M., and M. Yunus (2006). Serotype cycles in cholera dynamics. Proceedings of the Royal Society, B. 273. pp. 2879-2886.
Pascual, M., Koelle, K., and A. Dobson (2006). Hyperinfectivity in Cholera: A New Mechanism for an Old Epidemiological Model? PLoS Medicine. 3 (6). p. e280.
Koelle, K., Rodó, X., Pascual, M., Yunus, M., and G. Mostafa (2005). Refractory periods to climate forcing in cholera dynamics. Nature. 436. pp. 696-700.
Koelle, K., Pascual, M., and M. Yunus (2005). Pathogen adaptation to seasonal forcing and climate change. Proceedings of the Royal Society, B. 272. pp. 971-977.
Koelle, K. and J. Vandermeer (2005). Dispersal-induced desynchronization: from metapopulations to metacommunities. Ecology Letters. 8(2). pp. 167-175.
Buckee, C., Koelle, K., Mustard, M., and S. Gupta (2004). The effects of host contact network structure on pathogen diversity and strain structure. PNAS. 101(29). pp.10839-10844.
Koelle, K. and M. Pascual (2004). Disentangling extrinsic from intrinsic factors in disease dynamics: a nonlinear time series approach with an application to cholera. The American Naturalist 163(6), pp.901-913.
Savit, R., Koelle, K., Treynor, W., and R. Gonzalez (2004). Man and Superman: Human Limitations, Innovation, and Emergence in Resource Competition in Collectives and the Design of Complex Systems, eds. Tumer, K. & Wolpert, D. (Springer-Verlag, New York, NY).
Blower, S.M., Koelle, K., and J. Mills (2002). Health policy modeling: epidemic control, HIV vaccines and risky behavior in Quantitative Evaluation of HIV Prevention Programs, eds. Kaplan, E. H. & Brookmeyer, R. (Yale University Press, New Haven, CT).
Blower, S.M., Koelle, K., Kirschner, D.E., and J. Mills (2001). Live attenuated HIV vaccines: Predicting the tradeoff between efficacy and safety. PNAS. 98(6), pp. 3618-3623.
Blower, S.M., Koelle, K., and T. Lietman (1999). Antibiotic resistance- to treat… Nature Medicine 5(4), p. 358.