Current Capabilities
Protein Expression on Competent and Auxotrophic E. Coli cells (16 Liters)
Protein Synthesis using Microwave Peptide Synthesizer (CEM Liberty 1)
Site Directed Mutagenesis through PCR
Isotopic Labeling of Single Amino Acid or the Entire Protein
Protein Purification using FPLC and HPLC (Affinity Chromatography, Gel Filtration, and Reverse Phase Chromatography)
Protein Identification and Characterization using Non-Native Gels, UV-Vis Spectrometry, and Mass Spectrometry
Proteins are inherently noisy spectroscopic subjects, especially in the infrared. At times we need to be able to modify our samples to either clear up some of that noise or to create unique spectroscopic signals. We utilize the methionine auxotroph E. coli for some of our labeling. We express our proteins in a media lacking natural methionine. Instead we replace methionine with either 13C=18O backbone labeled methionine, shown as image b), or with the analog 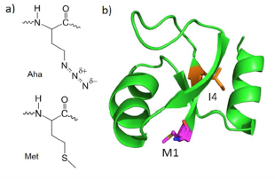 azidohomoalanine, shown as image a). The heavy backbone labeling shifts the amide I band lower in wavenumber so we can isolate the labeled residue’s C=O stretch. Azidohomoalanine has an azide group which produces an IR band around 2100 cm-1. This region is normally spectroscopically quiet and allows for a site specific label with high signal-to-noise. We also have the capability to express fully isotopically-labeled proteins. With these techniques, we are able to apply our various spectroscopic techniques to the whole protein or just specific residues.
azidohomoalanine, shown as image a). The heavy backbone labeling shifts the amide I band lower in wavenumber so we can isolate the labeled residue’s C=O stretch. Azidohomoalanine has an azide group which produces an IR band around 2100 cm-1. This region is normally spectroscopically quiet and allows for a site specific label with high signal-to-noise. We also have the capability to express fully isotopically-labeled proteins. With these techniques, we are able to apply our various spectroscopic techniques to the whole protein or just specific residues.

