Contributed by Runako Aranha-Minnis
Evolution is not only limited to the organisms we are able to see with the naked eye. Viruses like influenza are able to evolve, and this can be very dangerous to human populations. Though it is true that influenza is not the danger that it once was, it still can not be ignored. Viral resistance to treatment can lead to many deaths globally, especially during flu seasons. During these times, doctors usually see a rise in cases of viruses resistant to the treatments available at the time. One type of resistance that influenza can develop is Oseltamivir resistance. Oseltamivir is an important antiviral defense against the flu. It is found in drugs like Tamiflu, and can be rendered ineffective if resistance is developed.
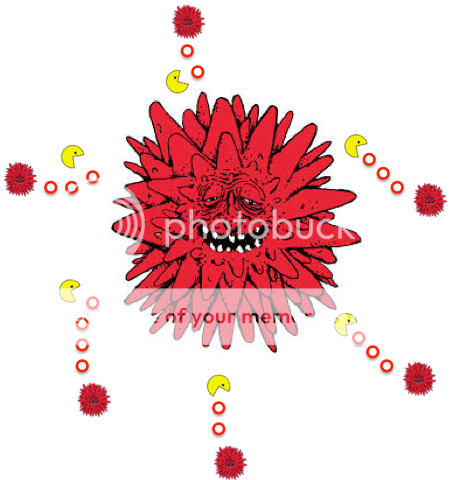
Influenza and its neuraminidase
The DNA change that in the virus that occurs is related to the neuraminidase enzyme activity. The enzyme cleaves salicylic acid moieties that can be bound by the viral hemagglutinin. In essence the enzyme’s job is to help release newly formed viral particles. Imagine the enzyme as little Pac-Men just going about and eating the connections of new virus particles to let them spread. Oseltamivir works by inhibiting the activity of the neuraminidase enzyme. This limits the infectiousness of the virus.
Resistance requires changes in the DNA of the virus. Mutations, changes in the DNA,  can change genes, often leading to changes in the organism. For Oseltamivir resistance, a point mutation, the change of a single base of DNA, is all that is necessary. A change in the genes leads to a change in the amino acids produced. This change in the amino acids is just one amino acid at the 274th spot in the chain leads to a change in the activity of the virus. This line of reasoning is why the mutation is called H274Y.
can change genes, often leading to changes in the organism. For Oseltamivir resistance, a point mutation, the change of a single base of DNA, is all that is necessary. A change in the genes leads to a change in the amino acids produced. This change in the amino acids is just one amino acid at the 274th spot in the chain leads to a change in the activity of the virus. This line of reasoning is why the mutation is called H274Y.
The H274Y mutation that gives influenza resistance usually does not spread through the population. In early clinical tests, the mutation meant the virus was unaffected by oseltamivir, but had associated decreased viral fitness. So even though viruses with resistance would be selected for, their lower chance of survivability because of the mutation meant it did not matter. The influenza virus is able to evolve with Oseltamivir only when an associated mutation occurs. These associated mutations that allow oseltamivir resistance to keep going are permissive mutations. More recently, permissive mutations have arisen that allow influenza to be resistant to Oseltamivir without losing any fitness. Imagine a population of influenza like a game of “Where is Waldo?”:
No permissive mutations. In the absence of permissive mutations, all viruses, including Waldo, die.
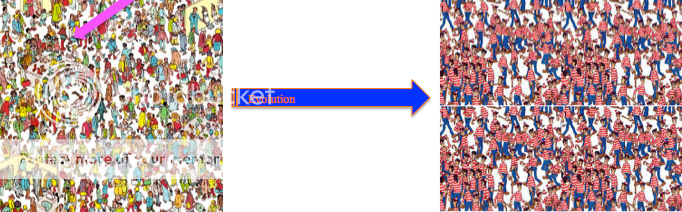
With permissive mutations. If Waldo has a permissive mutation, he survives to reproduce and the whole populations becomes Waldos because all other viruses died.
Waldo in the first cartoon has H274Y with no permissive mutations. Without permissive mutations, there is no evolution under treatment. After treatment, the viruses are killed. There are no viruses left so the picture goes black. In the second cartoon, With permissive mutations, there is evolution for Waldo and a new viral population arises. This population only has Waldo because only Waldo has the H274Y mutation that conferred resistance.
For Further Reading:
Jesse D. Bloom, Lizhi Ian Gong, David Baltimore. 2010. Permissive Secondary Mutations Enable the Evolution of Influenza Oseltamivir Resistance Science. 328: 1272-1275
Nicholas Renzette, Daniel R. Caffrey, Konstantin B. Zeldovich, Ping Liu, Glen R. Gallagher, Daniel Aiello, Alyssa J. Porter, Evelyn A. Kurt-Jones, Daniel N. Bolon, Yu-Ping Poh, Jeffrey D. Jensen, Celia A. Schiffer, Timothy F. Kowalik, Robert W. Finberg, Jennifer P. Wang. 2013. Evolution of the Influenza A Virus Genome during Development of Oseltamivir Resistance In Vitro 88: 272-281