Contributed by Ina Huang, Jenny Huang
Many people believe that natural selection subjectively selects the favorable traits in a population. But in fact, it is a passive process that does not involve organisms “trying” to adapt. Natural selection can be influenced by many factors, one of which is the random mutations that may occur. These mutations can range from those that influence the color adaptations we see in animals like the rat snake (Pantherophis obsoletus) to sickle cell disease in humans. Under different environmental conditions, some mutations lead to a higher survival rate of individuals and are therefore passed onto the next generation. This concept of the organism becoming more suited to its current environment is roughly the basis of adaptive evolution. This is a fundamental principle for natural selection instead of specific desires of species.
The generation of mutations in organisms is random. Nevertheless, there are attempts to comprehend the process of mutations. One emerging area of evolutionary biology that does this is called quantum biology. In quantum biology, the principle idea is that the interactions and movements of protons within the genome have a major influence on the entire organism. It is theorized that whether a mutation occurs or not in a gene is due to the position of a proton on that gene. The movement of such a small and basic component in DNA is able to generate drastic changes. It is speculated that shifts in the proton occur when the organism is exposed to selective conditions in their environment. These environmental factors induce selective pressures on the organism genes. Essentially, this means that at the level of the organism’s DNA, specific genes are placed under pressure. It is these pressures that provide the conditions that can induce mutation. First, it should be noted that whether or not the mutation occurs is random. However, like the organisms themselves, the protons at the atomic level are in constant motion. Thus mutations may arise at anytime.
 Let’s consider this idea with a simplified example using a strain of Escherichia coli that is nonfermenting. This means that this strain cannot use anaerobic respiration to obtain energy. If E.coli is grown on plates that are rich in lactose, a nutrient for growth, it has been shown that some colonies will develop a mutation that allows them to ferment or metabolize energy from lactose. So for those E.coli colonies on lactose-rich plates, to take advantage of the resources in their environment, a beneficial mutation can spontaneously occur that allows them to metabolize lactose. As mentioned previously, these mutations are random. They bacteria cannot force or influence themselves in anyway to gain the lactose-fermenting mutation. For the E. coli colonies that are able to utilize lactose and metabolize it, they are referred to as lac+. The strain that does not ferment on lactose-rich plates will be referred to as lac-.
Let’s consider this idea with a simplified example using a strain of Escherichia coli that is nonfermenting. This means that this strain cannot use anaerobic respiration to obtain energy. If E.coli is grown on plates that are rich in lactose, a nutrient for growth, it has been shown that some colonies will develop a mutation that allows them to ferment or metabolize energy from lactose. So for those E.coli colonies on lactose-rich plates, to take advantage of the resources in their environment, a beneficial mutation can spontaneously occur that allows them to metabolize lactose. As mentioned previously, these mutations are random. They bacteria cannot force or influence themselves in anyway to gain the lactose-fermenting mutation. For the E. coli colonies that are able to utilize lactose and metabolize it, they are referred to as lac+. The strain that does not ferment on lactose-rich plates will be referred to as lac-.
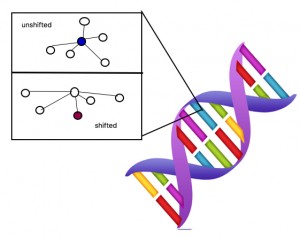
This figure shows a massive oversimplification of the atomic level in DNA. When the proton shifts the surroundings are also affected. This change is capable of forming the mutated gene.
Now, assume that there is a specific gene that controls the ability to ferment in E. coli. Like everything else on Earth, that gene is made up of protons, electrons, and neutrons at the atomic level. Let’s focus on one proton for now. If the proton is in shape A (AKA “configuration” in scientific language) then the E.coli is the wild type lac- , but if that proton shifts then it is lac+. The main idea is that the mutation in yeast is due to the change in the position of this one proton. Apparently, when the proton shifts this particular gene is pushed toward its mutated state. Though whether this shift occurs or not at some time period is random. When the environmental condition is providing some selective factor onto the genes of the organism the proton enters a state of decoherence, or instability. In this state the gene either crosses the line to become its mutated version or stays in the wild type form.
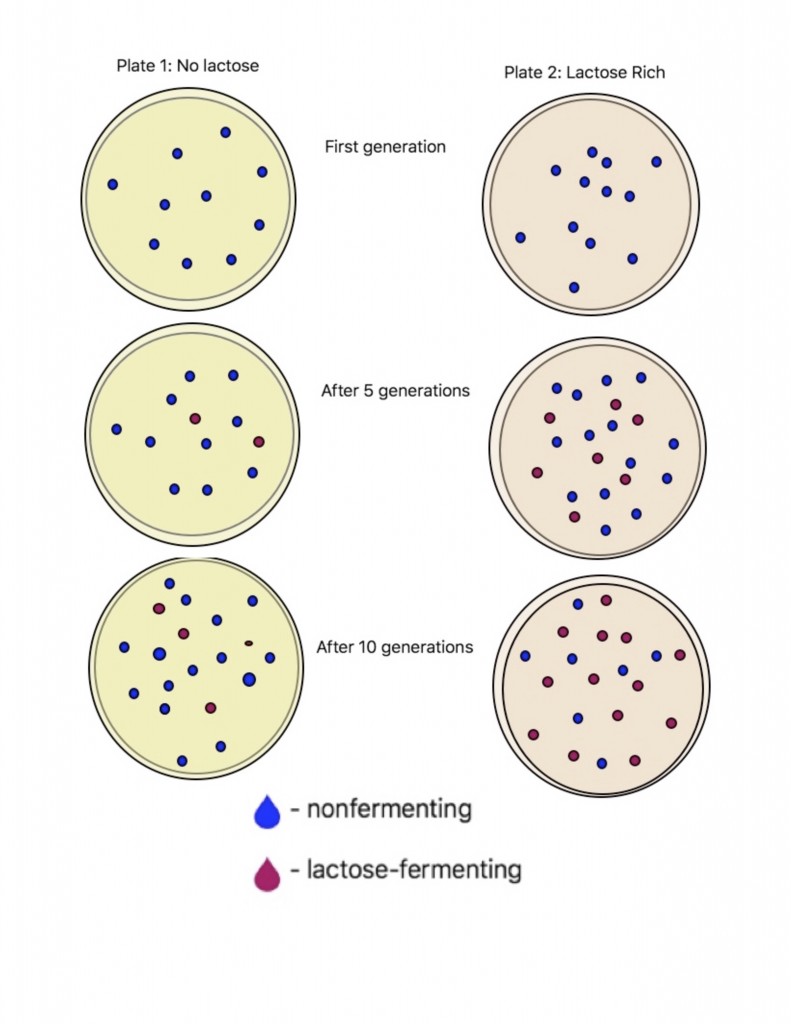
This images summaries that when E.coli is plated on lactose rich plates a greater percentage of the colonies become lac+. On the other hand, in the plates lacking lactose the occurrence of the lac+ mutation is less likely to occur.
Adaptive evolution is an interesting concept because it challenges the idea that mutations are completely random. As it is shown in the E. coli example, the environment impacted the gene under selection. The interactions with the environment in the example, allowed for the selection of lac+ E. coli. However, this does not mean that the organisms themselves have the abilities to influence which mutations may or may not arise. While, selective factors may direct the path of mutations in organisms, it does not induce the mutations. Individuals have no control over the formation of mutations. This area of adaptive mutation and the application of quantum biology is still relatively young. However, researching these two concepts provides more information on the evolutionary process.
Video Resource:
To learn more about how quantum biology relates to evolutionary biology watch the TEDtalk given by Jim Al-Khalili.
Reading resources for further information:
Bershtein, S., and D. S. Tawfik. 2008. ‘Ohno’s model revisited: measuring the frequency of potentially adaptive mutations under various mutational drifts’, Molecular Biology Evolution, 25: 2311-8.
Gerstein, A. C., D. S. Lo, and S. P. Otto. 2012. ‘Parallel genetic changes and nonparallel gene-environment interactions characterize the evolution of drug resistance in yeast’, Genetics, 192: 241-52.
Levin, B. R., and O. E. Cornejo. 2009. ‘The population and evolutionary dynamics of homologous gene recombination in bacterial populations’, PLoS Genet, 5: e1000601.
Lynch, M., and A. Abegg. 2010. ‘The rate of establishment of complex adaptations’, Molecular Biology Evolution 27: 1404-14.
McFadden, J., and J. Al-Khalili. 1999. ‘A quantum mechanical model of adaptive mutation’, Biosystems, 50: 203-11.
Ogryzko, V. 2009. ‘On two quantum approaches to adaptive mutations in bacteria’, NeuroQuantology, 7:564-595.