Contributed by Jonathan Nelson and Dina Michael
When Charles Darwin and Alfred Wallace presented their argument for speciation through natural selection, they established the foundations of evolutionary biology. Evolution is the process of change that lead from ape like ancestors to modern humans.
For data, scientist often turn to the fossil remains left by early hominids. Their remains exhibit a variety of species that likely evolved from Homo ergaster. Homo ergaster was a hominid species that possessed the capacity to disperse from Africa and colonize multiple areas of Europe and East Asia (Scarre 2005). These separated groups eventually underwent natural selection, and were likely influenced by genetic drift (certain alleles being randomly expressed and prevailing in the absence of natural selection). These developments lead to the evolution of the Homo sapien and Homo neanderthalensis species.
While many believe that modern humans emerged from these different hominid groups that were present throughout the world, modern evidence collected from mitochondrial DNA proves otherwise. Using this mitochondrial DNA, which is passed solely from mother to child, scientist postulate that all humans likely all came from one mother, who existed any where from 290,000 to 140,000 years ago (Cann 1987).
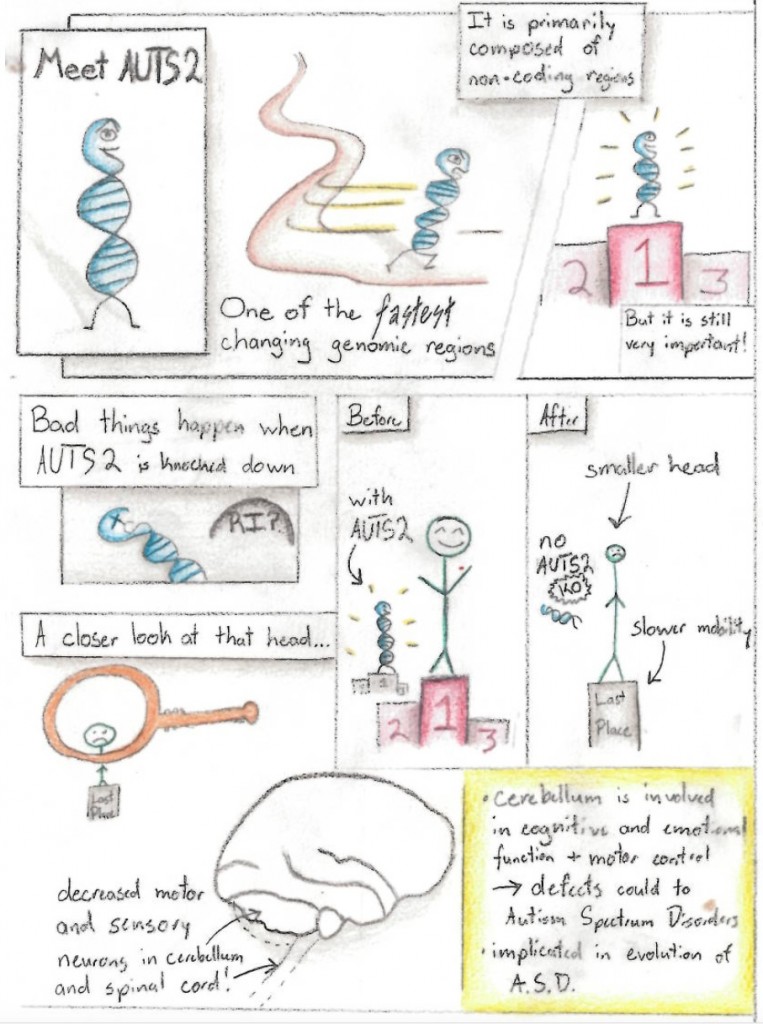
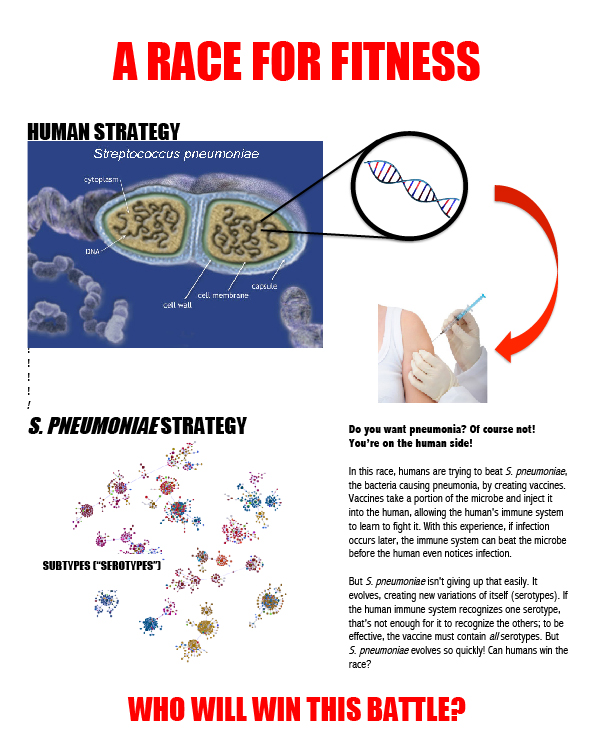
So why do we care? Well, it turns out that human evolution has plenty of modern implications as well. Modern research into human evolution and pathogens has shown that pathogens have played a large role in the shaping of human evolution. Researchers have found that certain pathogens in an area also drove the presence of certain alleles in a population, with the expression of genes such as those responsible for disorders like celiac disease and type 1 diabetes correlating with presence of certain pathogens in that area. They further hypothesized that traits evolved to survive in a pathogen rich environment may be the source of many autoimmune disorders, particularly in industrialized societies (Fumagalli 2011).
Mutation is one of the major driving forces of evolution,as it is the ultimate driving force behind variation. understanding the sources of these mutation is one major focuses of modern day research. In one study, scientist showed how the structure of DNA makes it very reactive with electromagnetic fields ( Blank and Goodman 2011) These researchers postulate that this may play a role in recent rises of cancer in our population. The malleability of our DNA only serves to show how we are continuously affected by the evolutionary process.
[jwplayer mediaid=”1296″]
For more information, please refer to the following sources:
Blank, M., & Goodman, R. 2011. DNA is a fractal antenna in electromagnetic fields. Int J Radiat Biol International Journal of Radiation Biology, 87(4), 409-415.
Cann, R. L., Stoneking, M., & Wilson, A. C.1987. Mitochondrial DNA and human evolution. Nature, 325(6099), 31-36.
Fumagalli, M., Sironi, M., Pozzoli, U., Ferrer-Admettla, A., Pattini, L., & Nielsen, R. 2011. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet, 7(11), e1002355.
Harding, R. M., Healy, E., Ray, A. J., Ellis, N. S., Flanagan, N., Todd, C., … Rees, J. L. (2000). Evidence for Variable Selective Pressures at MC1R. American Journal of Human Genetics, 66(4), 1351–1361.
Scarre, C.2005. The human past: World prehistory & the development of human societies . New York, NY: Thames & Hudson.pp. 85-99
Olson, M. V., & Varki, A. (2003). Sequencing the chimpanzee genome: insights into human evolution and disease. Nature Reviews Genetics, 4(1), 20-28.




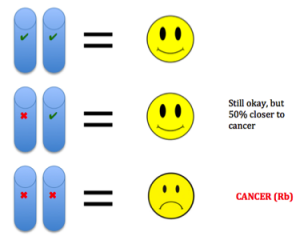 etic mutations and environmental exposures during one’s lifetime. Cancer is not heritable, but a predisposition to the disease can be (Peltomaki, 2012). For example, in our DNA we have two copies of each tumor suppressor gene, one from each parent. These genes keep cells from growing uncontrollably. So to lose function, one has to have mutations in both copies. Unfortunately, one can be born with a mutation in one copy, such as in Retinoblastoma (Rb). This means the gene is still expressed using the normal copy, but this confers a 50% increased predisposition for the disease bringing cells halfway closer to being cancer cells (Price et al., 2014).
etic mutations and environmental exposures during one’s lifetime. Cancer is not heritable, but a predisposition to the disease can be (Peltomaki, 2012). For example, in our DNA we have two copies of each tumor suppressor gene, one from each parent. These genes keep cells from growing uncontrollably. So to lose function, one has to have mutations in both copies. Unfortunately, one can be born with a mutation in one copy, such as in Retinoblastoma (Rb). This means the gene is still expressed using the normal copy, but this confers a 50% increased predisposition for the disease bringing cells halfway closer to being cancer cells (Price et al., 2014).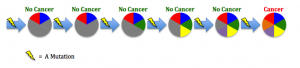 e different environments around cells fostering different mutations in various areas of the same tumor (Hardiman et al., 2015). Cancer is adapting to therapies that target mutations, making it so difficult to control.
e different environments around cells fostering different mutations in various areas of the same tumor (Hardiman et al., 2015). Cancer is adapting to therapies that target mutations, making it so difficult to control.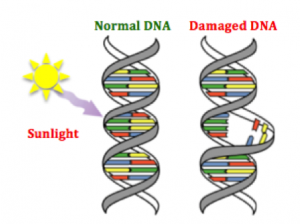 g agents are naturally occurring substances we encounter on a daily basis (Bauer, Corbett, & Doetsch, 2015). The human body has evolved ways to counteract DNA-damaging events, for example during sunlight exposure, using molecular machinery and likewise other naturally occurring compounds (Nishisgori, 2015).
g agents are naturally occurring substances we encounter on a daily basis (Bauer, Corbett, & Doetsch, 2015). The human body has evolved ways to counteract DNA-damaging events, for example during sunlight exposure, using molecular machinery and likewise other naturally occurring compounds (Nishisgori, 2015).